A straightforward technique to obtain genomic DNA from nasal swabs suitable for sheep SNP genotyping analysis
DOI:
https://doi.org/10.31285/AGRO.28.1452Keywords:
nasal swabs, ovine, DNA extraction, SNP array, concordance analysisAbstract
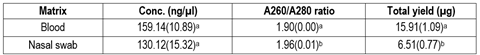
Isolation of high quality and quantity genomic DNA is essential for molecular studies. It is crucial to select a non-invasive and straightforward technique to ensure the efficient collection of DNA, particularly at the farm level. The aim of this study was to determine if nasal swabs are an appropriate biological matrix to obtain good quality genomic DNA suitable for SNP genotyping. In this study, two biological matrices (blood and nasal swabs) were evaluated and compared for the isolation of genomic DNA obtained from 15 female Texel sheep. DNA quality and quantity were assessed using spectrophotometry and gel electrophoreses. Genotype concordance rates were used for comparison. Results showed that the highest concentration mean was obtained from blood samples (159.14 ng/µl), while from nasal swab samples the concentration mean was lower (130.12 ng/µl), but the difference was non-significant. Regarding purity, DNA obtained from nasal swabs presented a higher A260/A280 ratio (1.96), while the one obtained from blood samples was 1.90. Total DNA yield obtained from blood samples (15.91 µg) was significantly higher than the one obtained from nasal swabs (6.51). Blood and nasal swab genotyping concordance rates were high (mean = 0.984). In conclusion, our results indicate that nasal swabs can yield good quality DNA; however, the DNA extraction protocol should be optimized.
Downloads
References
Aguilar I. SeekParentf90 [Internet]. Georgia: University of Georgia; 2017 [cited 2024 May 03]. Available from: http://nce.ads.uga.edu/wiki/doku.php?id=readme.seekparentf90
Chacon-Cortes D, Griffiths L. Methods for extracting genomic DNA from whole blood samples: current perspectives. Journal of Biorepository Science for Applied Medicine. 2014;2:1-9.Doi: 10.2147/BSAM.S46573. DOI: https://doi.org/10.2147/BSAM.S46573
Chiong KT, Damaj MB, Padilla CS, Avila CA, Pant SR, Mandadi KK, Ramos NR, Carvalho DV, Mirkov TE. Reproducible genomic DNA preparation from diverse crop species for molecular genetic applications. Plant Methods. 2017;13:106. Doi: 10.1186/s13007-017-0255-6. DOI: https://doi.org/10.1186/s13007-017-0255-6
Fan B, Du Z, Gorbach D, Rothschild M. Development and application of high-density SNP arrays in genomic studies of domestic animals. Anim Biosci. 2010;23(7):833-47. Doi:10.5713/ajas.2010.r.03. DOI: https://doi.org/10.5713/ajas.2010.r.03
Foley C, O'Farrelly C, Meade KG. Technical note: Comparative analyses of the quality and yield of genomic DNA from invasive and noninvasive, automated and manual extraction methods. J Dairy Sci. 2011;94(6):3159-65. Doi: 10.3168/jds.2010-3987. DOI: https://doi.org/10.3168/jds.2010-3987
Hawken RJ, Cavanagh JA, Meadows JR, Khatkar MS, Husaini Y, Zenger KR, McClintock S, McClintock AE, Raadsma HW. Technical note: Whole-genome amplification of DNA extracted from cattle semen samples. J Dairy Sci. 2006;89(6):2217-21. Doi: 10.3168/jds.S0022-0302(06)72292-5. DOI: https://doi.org/10.3168/jds.S0022-0302(06)72292-5
Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. Performance of Saliva, Oropharyngeal Swabs, and Nasal Swabs for SARS-CoV-2 Molecular Detection: a Systematic Review and Meta-analysis. J Clin Microbiol. 2021;59(5):e02881-20. Doi: 10.1128/JCM.02881-20. DOI: https://doi.org/10.1128/JCM.02881-20
Li G, Gelernter J, Kranzler HR, Zhao H. M(3): an improved SNP calling algorithm for Illumina BeadArray data. Bioinformatics. 2012;28(3):358-65. Doi: 10.1093/bioinformatics/btr673. DOI: https://doi.org/10.1093/bioinformatics/btr673
Masuda Y, Legarra A, Aguilar I, Misztal I. 331 Efficient quality control methods for genomic and pedigree data used in routine genomic evaluation. J Anim Sci. 2019;97(Suppl 3):50-1. Doi: 10.1093/jas/skz258.101. DOI: https://doi.org/10.1093/jas/skz258.101
Medrano JF, Aasen E, Sharrow L. DNA extraction from nucleated red blood cells. Biotechniques. 1990;8(1):43.
Murphy MA, Shariflou MR, Moran C. High quality genomic DNA extraction from large milk samples. J Dairy Res. 2002;69(4):645-9. Doi: 10.1017/s0022029902005848. DOI: https://doi.org/10.1017/S0022029902005848
Neary MT, Neary JM, Lund GK, Garry FB, Holt TN, Mohun TJ, Breckenridge RA. Technical note: A comparison of DNA collection methods in cattle and yaks. J Anim Sci. 2014;92(9):3811-5. Doi: 10.2527/jas.2013-7445. DOI: https://doi.org/10.2527/jas.2013-7445
Núñez L, Rodríguez MI, Giménez G, Martínez R. Evaluación y comparación de dos protocolos de extracción de ADN a partir de tres tipos de muestras de ovinos. Compend Cienc Vet. 2022;11(2):18-23. Doi: 10.18004/compend.cienc.vet.2021.11.02.18. DOI: https://doi.org/10.18004/compend.cienc.vet.2021.11.02.18
Psifidi A, Dovas CI, Banos G. A comparison of six methods for genomic DNA extraction suitable for PCR-based genotyping applications using ovine milk samples. Mol Cell Probes. 2010;24(2):93-8. Doi: 10.1016/j.mcp.2009.11.001. DOI: https://doi.org/10.1016/j.mcp.2009.11.001
Psifidi A, Dovas CI, Bramis G, Lazou T, Russel CL, Arsenos G, Banos G. Comparison of eleven methods for genomic DNA extraction suitable for large-scale whole-genome genotyping and long-term DNA banking using blood samples. PLoS One. 2015;10(1):e0115960. Doi: 10.1371/journal.pone.0115960. DOI: https://doi.org/10.1371/journal.pone.0115960
R Core Team, 2021. R: A language and environment for statistical computing [WWW Document]. R Found Stat Comput. URL https://www.R-project.org (accessed 4.29.22).
República Oriental del Uruguay, Poder Legislativo.Procedimientos para la utilizacion de animales en actividades de experimentacion, docencia e investigacion cientifica. Ley N° 18.611 [Internet]. 2009 [cited 2024 May 03]. Available from: https://www.impo.com.uy/bases/leyes/18611-2009
Sambrook J, Russell DW. The condensed protocols from molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2006. 800p. DOI: https://doi.org/10.1101/pdb.prot3919
Vignal A, Milan D, SanCristobal M, Eggen A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol. 2002;34(3):275-305. Doi: 10.1186/1297-9686-34-3-275. DOI: https://doi.org/10.1051/gse:2002009
Yang W, Kang X, Yang Q, Lin Y, Fang M. Review on the development of genotyping methods for assessing farm animal diversity. J Anim Sci Biotechnol. 2013;4(1):2. Doi: 10.1186/2049-1891-4-2. DOI: https://doi.org/10.1186/2049-1891-4-2
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Agrociencia Uruguay

This work is licensed under a Creative Commons Attribution 4.0 International License.
| Article metrics | |
|---|---|
| Abstract views | |
| Galley vies | |
| PDF Views | |
| HTML views | |
| Other views | |